MAIT is an R package to perform Metabolomic LC/MS analysis: MAIT (Metabolite Automatic Identification Toolkit), developed by Francesc Fernandez, Rafael Llorach, Cristina Andres-LaCueva and Alexandre Perera and it was published in Oxford Journals Bioinformatics (Fernández-Albert F., Llorach R., Andrés-Lacueva C., Perera-Lluna A. An R package to analyse LC/MS metabolomic data: MAIT (Metabolite Automatic Identification Toolkit). Bioinformatics (2014)).
 This software was jointly developed with University of Barcelona’s Nutrimetabolomics group, coordinated by Dra. Cristina Andres-Lacueva (candres@ub.edu).
This software was jointly developed with University of Barcelona’s Nutrimetabolomics group, coordinated by Dra. Cristina Andres-Lacueva (candres@ub.edu).
MAIT package is currently maintained by the B2SLab at UPC, so please send us a note if you have any suggestion or comment.
Please cite the following paper when using MAIT in your research:
MAIT Description
MAIT is capable of peak Peak detection for metabolomic LC/MS data sets. It uses a matched filter (Danielsson, Bylund, and Markides 2002) and the centWave algorithm (Tautenhahn et al. 2008) through the XCMS package, developed by the SCRIPPS Center for Metabolomics.
MAIT also uses further complementary steps in the peak annotation stage, also using CAMERA package (Kuhl et al. 2011). The peaks within each peak group are annotated following a reference adduct/fragment table and a mass tolerance window.
MAIT uses a predefined biotransformation table where the biotransformations to find are saved. A user-defined biotransformation table can be set as an input.
The package also holds a metabolite identification stage, in which a predefined metabolite database is mined to search for the significant masses, also using a tolerance window. This database is the Human Metabolome Database (HMDB) (Wishart, Knox, Guo, Eisner, Young, Gautam, Hau, Psychogios, Dong, Bouatra, and et al. 2009), 2009/07 version.
In terms of statistical tests, the package automatically tests on every feature and selects a significant set of features. Depending on the number of classes defined in the data, MAIT can use Student’s T-test, Welch’s T-tests and Mann-Whitney tests for two classes or ANOVA and Kruskal-Wallis tests for more than two classes. The use is able to define custom tests in the automatic procedure as well.
A nice addition of the MAIT R package, is the support for peak aggregation techniques that might lead to better feature selection (Fernández-Albert, Llorach, Andrés-Lacueva, and Perera-Lluna), as defined in:
Downloading MAIT
The latest version of the MAIT package is available through the Bioconductor repository:
- Package: MAIT
- Tutorial: MAITSupplementary
- Manual: MAIT Manual
Data of the faahKO package in zip format. This is the data used in the MAIT tutorial.
Using MAIT
The data files for this example are a subset of the data used in reference (Saghatelian, Trauger, Want, Hawkins, Siuzdak, and Cravatt 2004), which are freely distributed through the faahKO package Smith (2012). In these data there are 2 classes of mice: a group where the fatty acid amide hydrolase gene has been suppressed (class knockout or KO) and a group of wild type mice (class wild type or WT). There are 6 spinal cord samples in each class. In the following, the MAIT package will be used to read and analyse these samples using the main functions discussed in Section 5. The significant features related to each class will be found using statistical tests and analysed through the different plots that MAIT produces.
Data Import
Each sample class file should be placed in a directory with the class name. All the class folders should be placed under a directory containing only the folders with the files to be analysed. In this case, 2 classes are present in the data. An example of correct file distribution using the example data files is shown in the following figure.
Figure 2: Example of the correct sample distribution for MAIT package use. Each sample file has to be saved under a folder with its class name.
Peak Detection
Once the data is placed in 2 subdirectories of a single folder, the function sampleProcessing() is run to detect the peaks, group the peaks across samples, perform the retention time correction and carry out the peak filling process. As function sampleProcessing() uses
the XCMS package to perform these 4 processing steps, this function exposes XCMS parameters that might be modified to improve the peak detection step. A project name should be defined because all the tables and plots will be saved in a folder using that name. For example, typing project = “project_Test”, the output result folder will be “Results_project_Test”. By choosing “MAIT_Demo” as the project name, the peak detection stage can be launched by typing:
R> MAIT <- sampleProcessing(dataDir= "Dataxcms", project = "MAIT_Demo", snThres=2,rtStep = 0.03) ko15: 215:366 230:680 245:1014 260:1392 275:1766 290:2120 305:2468 320:2804 335:3150 350:3468 365:3846 380:4182 395:4486 410:4804 425:5110 440:5444 455:5778 470:6136 485:6504 500:6892 515:7296 530:7742 545:8138 560:8620 575:9048 590:9526 . . . Peak detection done 262 325 387 450 512 575 Retention Time Correction Groups: 7 Warning: Span too small, resetting to 0.8 Retention time correction done 262 325 387 450 512 575 Peak grouping after samples done ko15 . . . Missing Peak integration done
After having launched the sampleProcessing function, peaks are detected, they are grouped across samples and their retention time values are corrected. A short summary in the R session can be retrieved by typing the name of the MAIT-class object.
R> MAIT A MAIT object built of 12 samples The object contains 6 samples of class KO The object contains 6 samples of class WT
The result is a MAIT-class object that contains information about the peaks detected, their class names and how many files each class contains. A longer summary of the data is retrieved by performing a summary of a MAIT-class object. In this longer summary version, further information related to the input parameters of the whole analysis is displayed. This functionality is especially useful in terms of traceability of the analysis.
R> summary(MAIT)
A MAIT object built of 12 samples
The object contains 6 samples of class KO
The object contains 6 samples of class WT
Parameters of the analysis:
Value
dataDir "Data"
snThres "2"
Sigma "2.123"
mzSlices "0.3"
retcorrMethod "loess"
groupMethod "density"
bwGroup "3"
mzWidGroup "0.25"
filterMethod "matchedFilter"
rtStep "0.03"
nSlaves "0"
project "MAIT_Demo"
ppm "10"
minfrac "0.5"
fwhm "30"
family1 "gaussian"
family2 "symmetric"
span "0.2"
centWave peakWidth1 "5"
centWave peakWidth2 "20"
Peak Annotation
The next step in the data processing is the first peak annotation step, which is per- formed through the peakAnnotation(). If the input parameter adductTable is not set, then the default MAIT table for positive polarisation will be selected. However, if the adductTable parameter is set to ”negAdducts”, the default MAIT table for negative fragments will be chosen instead. peakAnnotation function also creates an output table (see Table 3) containing the peak mass (in charge/mass units), the retention time (in minutes) and the spectral ID number for all the peaks detected. A call of the function peakAnnotation may be:
R> MAIT <- peakAnnotation(MAIT.object = MAIT, corrBetSamp = 0.75, perfwhm = 0.6) WARNING: No input adduct/fragment table was given. Selectinf default MAIT table for positive polarity Set adductTable equal to negAdducts to use the default MAIT table for negative polarity Start grouping after retention time. Created 1037 pseudospectra Spectrum build after retention time done Generating peak matrix! Run isotope peak annotation %finished: 10 20 30 40 50 60 70 80 90 100 Found isotopes: 15 Isotope Annotation done Start grouping after correlation Generating EIC Calculating peak correlations in 1037 groups... %finished: 10 20 30 40 50 60 70 80 90 100 Calculating peak correlations across samples %finished: 10 20 30 40 50 60 70 80 90 100 Calculate isotope assignments in 1037 Groups... %finished: 10 20 30 40 50 60 70 80 90 100 Calculating graph cross linking in 1037 groups... %finished: 10 20 30 40 50 60 70 80 90 100 New number of ps-groups: 2398 xsAnnotate has now 2398 groups, instead of 1037 Spectrum number increase after correlation done. Generating peak matrix for peak annotation! Found and use user-defined ruleset! Calculating possible adducts in 2398 Groups... %finished: 10 20 30 40 50 60 70 80 90 100 Adduct/fragment annotation done
Because the parameter adductTable was not set in the peakAnnotation call, a warning was shown informing that the default MAIT table for positive polarisation mode was selected. The xsAnnotated object that contains all the information related to peaks, spectra and their annotation is stored in the MAIT object. It can be retrieved by typing:
R> rawData(MAIT) $xsaFA An "xsAnnotate" object! With 2398 groups (pseudospectra) With 12 samples and 2640 peaks Polarity mode is set to: positive Using automatic sample selection Annotated isotopes: 15 Annotated adducts & fragments: 16 Memory usage: 7.07 MB
Following the first peak annotation stage, we want to know which features are different be- tween classes. Consequently, we run the function spectralSigFeatures().
R> MAIT<-spectralSigFeatures(MAIT.object = MAIT, pvalue = 0.05, p.adj = "none", scale = FALSE)
It is worth mentioning that by setting the scale parameter to TRUE, the data will be scaled to have unit variance. The parameter p.adj allows for using the multiple testing correction methods included in the function p.adjust of the package stats. A summary of the statis- tically significant features is created and saved in a table called significantFeatures.csv (see Table 3). It is placed inside the Tables subfolder located in the project folder. This table shows characteristics of the statistically significant features, such as their P-value, the peak annotation or the expression of the peaks across samples. This table can be retrieved at any time from the MAIT-class objects by typing the instruction:
R> signTable <- sigPeaksTable(MAIT.object = MAIT, printCSVfile = FALSE) R> head(signTable) mz mzmin mzmax rt rtmin rtmax npeaks KO WT ko15 ... 610 300.2 300.1 300.2 56.36 56.18 56.56 17 6 3 4005711.4 ... 762 326.2 326.1 326.2 56.92 56.79 57.00 9 5 2 3184086.4 ... 885 348.2 348.1 348.2 56.95 56.79 57.15 14 4 2 320468.2 ... 1760 495.3 495.2 495.3 56.93 56.82 57.05 11 3 4 110811.4 ... 935 356.2 356.1 356.3 63.77 63.58 63.92 9 4 4 962224.6 ... 1259 412.2 412.1 412.3 68.61 68.44 68.81 16 4 3 113096.3 ... isotopes adduct pcgroup P.adjust p 610 27 0.01748294 0.01748294 ... 762 [M+H]+ 325.202 31 0.01991433 0.01991433 ... 885 [M+Na]+ 325.202 31 0.16856322 0.16856322 ... 1760 31 0.96828618 0.96828618 ... 935 74 0.03310409 0.03310409 ... 1259 81 0.02240898 0.2240898 ... Median Class KO Median Class WT 610 2769931.356 115642.29 762 2353947.791 43006.61 885 40384.825 0.00 1760 6531.515 15969.26 935 848999.980 16836.67 1259 215979.768 34607.95
The number of significant features can be retrieved from the MAIT-class object as follows:
R> MAIT A MAIT object built of 12 samples and 2640 peaks. No peak aggregation technique has been applied 106 of these peaks are statistically significant The object contains 6 samples of class KO The object contains 6 samples of class WT
By default, when using two classes, the statistical test applied by MAIT is the Welch’s test. Nevertheless, when having two classes,MAIT also supports applying the Student’s t-test and the non-parametric test Mann-Whitney test. For using the Student’s t-test on the data, the call to the spectralSigFeatures function should be as:
R> MAIT_Student <- spectralSigFeatures(MAIT.object = MAIT, pvalue = 0.05, p.adj = "none", scale = FALSE, var.equal = TRUE) R> MAIT_Student A MAIT object built of 12 samples and 2640 peaks.
No peak aggregation technique has been applied 148 of these peaks are statistically significant The object contains 6 samples of class KO The object contains 6 samples of class WT
If we want to apply the Mann-Whitney test, in this case is necessary to add some jitter noise in our data. The reason is that the Mann-Whitney test has ties when the data has values equal to zero. Adding a small noise to the data solves this issue. MAIT supports using jitter noise through the flag jitter and the parameter jitter.amount:
R> MAIT_MW <- spectralSigFeatures(MAIT.object = MAIT, pvalue = 0.05, p.adj = "none", scale = FALSE, parametric = FALSE, jitter = TRUE)
As an example of its modularity, MAIT supports applying user-defined statistical tests on the data. In the following example, lets suppose that we want to compute the p-values corresponding to the exact Fisher’s test. To use a user-defined test in MAIT, the output of the function should give the p-value of the test given a numeric vector (i.e. the variable values) and a factor vector (the classes of the samples in the parameter group). In our case, the function of the Fisher’s test, can be defined as follows:
R> ftest <- function(x,group){return(fisher.test(x=x,y=group)$p.value)}
And the call to the spectralSigFeatures in this case:
R> MAIT_Fisher <- spectralSigFeatures(MAIT.object = MAIT, test.fun = ftest, namefun = "fisher's test") R> MAIT_Fisher A MAIT object built of 12 samples and 2640 peaks. No peak aggregation technique has been applied 18 of these peaks are statistically significant The object contains 6 samples of class KO The object contains 6 samples of class WT
The argument test.fun contains the function of the user-defined test and the argument namefun is an optional parameter that contains the name of the user-defined function. This name will appear in the parameters slot of the MAIT-class object:
R> summary(MAIT_Fisher)
A MAIT object built of 12 samples and 2640 peaks.
No peak aggregation technique has been applied
18 of these peaks are statistically significant
The object contains 6 samples of class KO
The object contains 6 samples of class WT
Parameters of the analysis
Value
dataDir "Data"
snThres "2.123"
mzSlices "0.3"
retcorrMethod "loess"
groupMethod "density"
bwGroup "3"
mzWidGroup "0.25"
filterMethod "matchedFilter"
rtStep "0.03"
nSlaves "0"
project "MAIT_Demo"
ppm "10"
minfrac "0.5"
fwhm "30"
family1 "gaussian"
family2 "symmetric"
span "0.2"
centWave peakwidth1 "5"
centWave peakwidth2 "20"
corrWithSamp "0.7"
corrBetSamp "0.75"
perfwhm "0.6"
sigma "6"
peakAnnotation pvalue "0.05"
calcIso "TRUE"
calcCiS "TRUE"
calcCaS "TRUE"
graphMethod "hcs"
annotateAdducts "TRUE"
peakAggregationMethod "None"
peakAggregation PCAscale "FALSE"
peakAggregation PCAcenter "FALSE"
peakAggregation scale "FALSE"
peakAggregation RemoveOnePeakSpectra "FALSE"
fisher's test pvalue "0.05"
fisher's test p-value p.adj "none"
The multiple test corrections are also implemented in this case by changing the p.adj argument of the function:
R> MAIT_Fisher <- spectralSigFeatures(MAIT.object = MAIT, test.fun = ftest, namefun = "fisher's test", p.adj = "fdr") Warning message: In spectralSigFeatures(MAIT.object = MAIT, test.fun = ftest, namefun ="fisher's test", : No significative features found with the selected parameters.
In this particular case, a warning is thrown as no significant features were found with a false discovery rate-adjusted p-value lower or equal 0.05.
Statistical Plots
Out of 2,402 features, 106 were found to be statistically significant. At this point, several MAIT functions can be used to extract and visualise the results of the analysis. Functions plotBoxplot, plotHeatmap, plotPCA and plotPLS automatically generate boxplots, heat maps PCA score plot and PLS score plot files in the project folder when they are applied to a MAIT object (see Table 3).
R> plotBoxplot(MAIT) R> plotHeatmap(MAIT) R> MAIT<-plotPCA(MAIT) R> MAIT<-plotPLS(MAIT)
The plotPCA and plotPLS functions produce MAIT objects with the corresponding PCA and PLS models saved inside. The models, loadings and scores can be retrieved from the MAIT objects by using the functions model, loadings and scores:
R> PLSmodel <- model(x=MAIT, type = "PLS")
R> PCAmodel <- model(x=MAIT, type = "PCA")
R> PLSscores <- scores(x=MAIT,model="PLS")
R> PCAscores <- scores(x=MAIT,model="PCA")
R> PLSloadings <- loadings(x=MAIT,model="PLS")
R> PCAloadings <- loadings(x=MAIT,model="PCA")
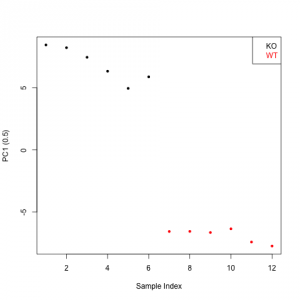
R> PLSscores
Comp1
1 8.460117
2 8.238226
3 7.465394
4 6.341839
5 4.958885
6 5.887925
7 -6.577803
8 -6.570983
9 -6.660059
10 -6.363424
11 -7.427228
12 -7.752889
attr(,"class")
[1] "scores"
R> PCAscores[,1:3]
PC1 PC2 PC3
[1,] -8.758728 0.92480221 -6.1406083
[2,] -8.348530 -0.86569846 0.1783953
[3,] -7.570347 0.32825445 -1.6159867
[4,] -6.209758 -0.01281555 3.1104855
[5,] -4.632576 -0.80459247 5.6779015
[6,] -5.757966 -0.47710433 0.8561668
[7,] 6.483476 7.10158291 0.9827710
[8,] 6.508645 0.44504996 -1.2287543
[9,] 6.568818 3.66149693 -0.2422269
[10,] 6.311563 -1.97819990 -0.8625683
[11,] 7.518147 -5.26076372 -0.8812214
[12,] 7.887257 -3.06201203 0.1656458
R> head(matrix(PLSloadings))
[,1]
[1,] 0.11179158
[2,] 0.10718688
[3,] 0.10167223
[4,] 0.10124325
[5,] -0.09481443
[6,] 0.10828112
R> head(PCAloadings[,1:3])
PC1 PC2 PC3
[1,] -0.1129682 0.008376894 -0.14442144
[2,] -0.1080615 -0.002674411 -0.14786276
[3,] -0.1027608 -0.006700719 -0.10304058
[4,] -0.1009138 -0.010796632 0.09038020
[5,] 0.0950440 -0.212358347 -0.06243794
[6,] -0.1098603 0.054060752 -0.16588612
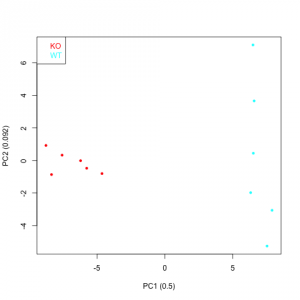
All the output figures are saved in their corresponding subfolders contained in the project folder. The names of the folders for the boxplots, heat maps and score plots are Boxplots, Heatmaps, PCA Scoreplots and PLS Scoreplots respectively. Figures 3 and 4 depict a heat map, a PCA score plot and a PLS score plot created when functions plotHeatmap, plotPCA and plotPLS were launched. Inside the R session, the project folder is recovered by typing:
R> resultsPath(MAIT)
Biotransformations
Before identifying the metabolites, peak annotation can be improved using the function Biotransformations to make interpreting the results easier. The MAIT package uses a default biotransformations table, but another table can be defined by the user and introduced by using the bioTable function input variable. The biotransformations table that MAIT uses is saved inside the file MAITtables.RData, under the name biotransformationsTable.
R> MAIT <- Biotransformations(MAIT.object = MAIT, peakPrecision = 0.005, adductAnnotation=FALSE) WARNING: No input biotransformations table was given. Selecting default MAIT table for biotransformations... WARNING: No input adduct/fragment table was given. Selecting default MAIT table for positive polarity... Set adductTable equal to negAdducts to use the default MAIT table for negative polarity % Annotation in progress: 10 20 30 40 60 70 80 90 100
The Biotransformations function can also annotate adducts by setting the flag adductAnnotation as TRUE. This is useful when analysing peak data that come from an external source (i.e. peaks and spectra have not been detected by MAIT).
Building a user-defined biotransformations table from the MAIT default table or adding a new biotransformation is straightforward. For example, let’s say we want to add a new adduct called ”custom biotrans” whose mass loss is 105.
R> data(MAITtables)
R> myBiotransformation<-c("custom_biotrans",105.0)
R> myBiotable<-biotransformationsTable
R> myBiotable[,1]<-as.character(myBiotable[,1])
R> myBiotable<-rbind(myBiotable,myBiotransformation)
R> myBiotable[,1]<-as.factor(myBiotable[,1])
R> tail(myBiotable)
NAME MASSDIFF 45 glucuronide conjugation 176.0321 46 hydroxylation + glucuronide 192.0270 47 GSH conjugation 305.0682 48 2x glucuronide conjugation 352.0642 49 [C13] 1.0034 50 custom_biotrans 105.0
Figure 3: Heat map created by the function plotHeatmap. Row numbers refer to spectra numbers.
Figure 4: PCA and PLS score plots (top and bottom plots respectively) generated by functions plotPCA and plotPLS. The PLS decomposition in this case has just one principal component.
To build an entire new biotransformations table, you only need to follow the format of the biotransformationsTable, which means writing the name of the biotransformations as factors in the NAME field of the data frame and their corresponding mass losses in the MASSDIFF field.
Metabolite Identification
Once the biotransformations annotation step is finished, the significant features have been enriched with a more specific annotation. The annotation procedure performed by the Biotransformations() function never replaces the peak annotations already done by other functions. MAIT considers the peak annotations to be complementary; therefore, when new annotations are detected, they are added to the current peak annotation and the identifica- tion function may be launched to identify the metabolites corresponding to the statistically significant features in the data.
R> MAIT <- identifyMetabolites(MAIT.object = MAIT, peakTolerance = 0.005, polarity="positive")
R> MAIT <- identifyMetabolites(MAIT.object = MAIT, peakTolerance = 0.005, polarity="positive") WARNING: No input database table was given. Selecting default MAIT database... Metabolite identification initiated % Metabolite identification in progress: 10 20 30 40 50 60 70 80 90 100 Metabolite identification finished
By default, the function identifyMetabolites() looks for the peaks of the significant fea- tures in the MAIT default metabolite database. The input parameter peakTolerance defines the tolerance between the peak and a database compound to be considered a possible match. It is set to 0.005 mass/charge units by default. The argument polarity, refers to to the polar- ity in which the samples were taken (positive or negative). It is set to “positive” by default but it should be adjusted changed to ”negative” if the samples were recorded in negative po- larisation mode. To check the results easily, function identifyMetabolites creates a table containing the significant feature characteristics and the possible metabolite identifications. Such a table is recovered from the MAIT-class object using the instruction:
R> metTable <- metaboliteTable(MAIT) R> head(metTable)
This table provides useful results about the analysis of the samples, such as the P-value of the statistical test, its adduct or isotope annotation and the name of any possible hit in the database. Note that if no metabolite has been found in the database for a certain feature, it is labelled as “unknown” in the table. The table also includes the median and mean values per class and feature.
Validation
Finally, we will use the function Validation() to check the predictive value of the significant features. All the information related to the output of the Validation() function is saved in the project directory in a folder called “Validation”. Two boxplots showing the overall and per class classification ratios are created along with a confusion matrix corresponding for each iteration (see Table 3).
R> MAIT <- Validation(Iterations = 20, trainSamples= 3, MAIT.object = MAIT)
Iteration 1 done Iteration 2 done Iteration 3 done
…
Iteration 19 done Iteration 20 done
A summary of a MAIT object, which includes the overall classification values, can be accessed:
R> summary(MAIT)
A MAIT object built of 12 samples and 2640 peaks. No peak aggregation technique has been applied
106 of these peaks are statistically significant
The object contains 6 samples of class KO
The object contains 6 samples of class WT
The Classification using 3 training samples and 20 Iterations gave the results:
KNN PLSDA SVM
mean 1 1 1
standard error 0 0 0
Parameters of the analysis:
.
.
.
It is also possible to gather the classification ratios per class, classifier used and iteration number by using the function classifRatioClasses():
R> classifRatioClasses(MAIT)
The classification ratios are 100% in all the iterations; the set of significant features separates the samples belonging to these classes.
Using External Peak Data
Taking advantage of the modularised design of MAIT, it is possible to use the function MAITbuilder to import peak data and analyse it using the MAIT statistical functions. As stated in section 5.5, there are certain arguments that should be provided depending on which function is wanted to be launched. In this section we will show an example of this data importation procedure using the same data that we have been using in the tutorial so far. Let’s say we have a peak table recorded in positive polarisation mode with the peak masses and retention time values such as:
R> peaks <- scores(MAIT) R> masses <- getPeaklist(MAIT)$mz R> rt <- getPeaklist(MAIT)$rt/60
We want to perform an annotation stage and metabolite identification on these data. To that end, we can launch the function MAITbuilder to build a MAIT-class object with the data in the table:
R> importMAIT <- MAITbuilder(data = peaks, masses = masses, rt = rt,
significantFeatures = TRUE, spectraEstimation = TRUE, rtRange=0.2,corThresh=0.7)
R> peaks <- scores(MAIT) R> importMAIT <- MAITbuilder(data = peaks, masses = masses, rt = rt, significantFeatures = TRUE, spectraEstimation = TRUE, rtRange=0.2, corThresh=0.7)
We have selected the option spectraEstimation as TRUE because we do not know the grouping of the peaks into spectra. As we want to annotate and identify all the peaks in the data frame, we set the flag significantFeatures to TRUE. At this point, we can launch the Biotransformations function:
R> importMAIT <- Biotransformations(MAIT.object = importMAIT, adductAnnotation = TRUE, peakPrecision = 0.005, adductTable=NULL)
We set the adductAnnotation flag to TRUE as we want to perform an adduct annotation step. The parameter adductTable set to NULL implies that a positive polarisation adduct annotation stage will be performed. To run a negative annotation, the argument should be set to negAdducts. The metabolite identification stage is launched as in the previous case:
R> importMAIT <- identifyMetabolites(MAIT.object = importMAIT, peakTolerance=0.005, polarity="positive")
The annotation of the Biotransformations and the adducts is given in the Adduct field of the metabo- lite table. The identification procedure can be performed for LC/MS data gathered in negative po- larisation mode by setting polarity = “negative”. If the class information is also introduced in the MAITbuilder, it is also possible to launch the computation of statistical tests (through function spectralSigFeatures), the validation and the functions regarding the statistical plots and models.

Pingback: MAIT package now available in Bioconductor! - B2SLab