Thanks to the highly reproducible ionization process of electron ionization (EI), gas chromatography coupled to mass spectrometry (GC/MS) has been a long-standing approach used for identifying small molecules. However, this approach in untargeted metabolomics produces large and complex data sets, characterized by coeluting compounds and extensive fragmentation of molecular ions caused by the hard electron ionization. In order to identify and extract quantitative information on metabolites across multiple biological samples, integrated computational workflows for data processing are needed.
 The Metabolomics Platform, Department of Electronic Engineering (DEEEA), Universitat Rovira i Virgili, Institut de Recerca Pediàtrica, Hospital Sant Joan de Déu, and Bioinformatics and Biomedical Signals Laboratory (B2SLab) from Center for Biomedical Engineering Research (CREB) announce eRah, a free computational tool conceived to fulfil this necessity.
The Metabolomics Platform, Department of Electronic Engineering (DEEEA), Universitat Rovira i Virgili, Institut de Recerca Pediàtrica, Hospital Sant Joan de Déu, and Bioinformatics and Biomedical Signals Laboratory (B2SLab) from Center for Biomedical Engineering Research (CREB) announce eRah, a free computational tool conceived to fulfil this necessity.
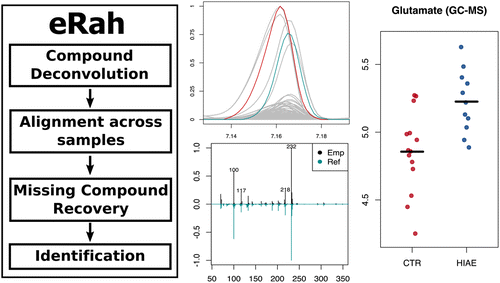
It is written in the open language R and it is composed of five core functions: (i) noise filtering and baseline removal of GC/MS chromatograms, (ii) an innovative compound deconvolution process using multivariate analysis techniques based on compound match by local covariance (CMLC) and orthogonal signal deconvolution (OSD), (iii) alignment of mass spectra across samples, (iv) missing compound recovery, and (v) identification of metabolites by spectral library matching using publicly available mass spectra. eRah outputs a table with compound names, matching scores and the integrated area of compounds for each sample.
The automated capabilities of eRah are demonstrated by the analysis of GC-time-of-flight (TOF) MS data from plasma samples of adolescents with hyperinsulinaemic androgen excess and healthy controls. The quantitative results of eRah are compared to centWave, the peak-picking algorithm implemented in the widely used XCMS package, MetAlign, and ChromaTOF software. Significantly dysregulated metabolites are further validated using pure standards and targeted analysis by GC-triple quadrupole (QqQ) MS, LC-QqQ, and NMR.
eRah is freely available at http://CRAN.R-project.org/package=erah.




Reference:
Xavier Domingo-Almenara, Jesus Brezmes, Maria Vinaixa, Sara Samino, Noelia Ramirez, Marta Ramon-Krauel, Carles Lerin, Marta Díaz, Lourdes Ibáñez, Xavier Correig, Alexandre Perera-Lluna, and Oscar Yanes (2016). eRah: A Computational Tool Integrating Spectral Deconvolution and Alignment with Quantification and Identification of Metabolites in GC/MS-Based Metabolomics. Anal. Chem., 88 (19), pp 9821–9829.
